I am a molecular biologist which means that I study the organization and functions of the genes and proteins that make up cells. A major flaw in my field is that we tend to be obsessive about the minutest details. Whole PhD projects can focus on the role of one gene (among thousands). Why is this work important? It helps reveal the innermost mechanism of a cell. These cells include dangerous bacteria, such as the ones that cause tuberculosis (TB).
Within a cell, genes do not function in isolation. They work in pathways that are dependent on one another. This means, for 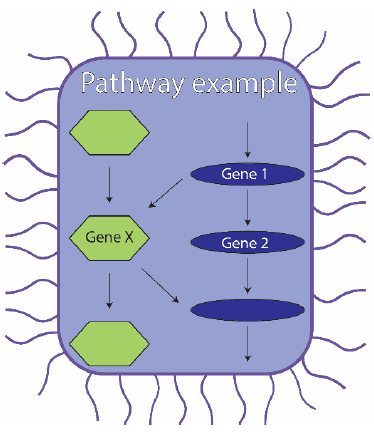 example, that in the blue pathway illustrated in the figure, the function of gene 2 is dependent on gene 1’s function. Gene X is part of a separate (green) pathway (in the same cell) which has its own function. However, in the absence of gene 2, gene X can substitute for gene 2 even though they normally function in different pathways (like a soccer player substituting for a basketball player). Sound complicated? That’s because it is. So it’s often easier to focus on only one gene. But then you won’t notice any substitution in the pathway. Back to our example, this means that we won’t be aware that gene 2 and gene X can have the same function. This is what my project is studying. We want to look at many different pathways to see if we can understand how they interact. To do this we will be building a library. Not the kind with books, but a library of cells, called a CRISPR interference library.
example, that in the blue pathway illustrated in the figure, the function of gene 2 is dependent on gene 1’s function. Gene X is part of a separate (green) pathway (in the same cell) which has its own function. However, in the absence of gene 2, gene X can substitute for gene 2 even though they normally function in different pathways (like a soccer player substituting for a basketball player). Sound complicated? That’s because it is. So it’s often easier to focus on only one gene. But then you won’t notice any substitution in the pathway. Back to our example, this means that we won’t be aware that gene 2 and gene X can have the same function. This is what my project is studying. We want to look at many different pathways to see if we can understand how they interact. To do this we will be building a library. Not the kind with books, but a library of cells, called a CRISPR interference library.
First, let me explain the basics. CRISPR is a revolutionary new technique that allows for the precise cleavage of DNA sequences. CRISPR interference (CRISPRi) is a modified version of this technique that allows us to switch genes off – this means that the target genes do not synthesize their usual products. The absence of some gene products will cause the cell to die, while others are not required for survival. To try and understand which genes need to be switched off to cause cell death, researchers must find ways of looking at lots of different cells with different switched off genes at the same time. Scaling up this process is called making a library. Think of each “book” as a cell and the “message” in each book the instruction to switch off a particular, individual gene. CRISPRi is the tool used to switch off the gene. Importantly, our biological library has a cataloguing system. After activating CRISPRi, the researcher checks which books (cells) are missing from the library and the missing catalogue numbers tells them which gene, when switched off, cause cell death. Because we are interested in identifying substitutions in pathways, our library will be switching off two genes at once. That means that each “book” or cell will contain two “messages” telling CRISPRi to switch-off two different genes in the same cell. Because we want to look at every possible gene pair of 140 genes, our library will contain 19 600 unique “messages”. That’s a lot of books to keep track of.
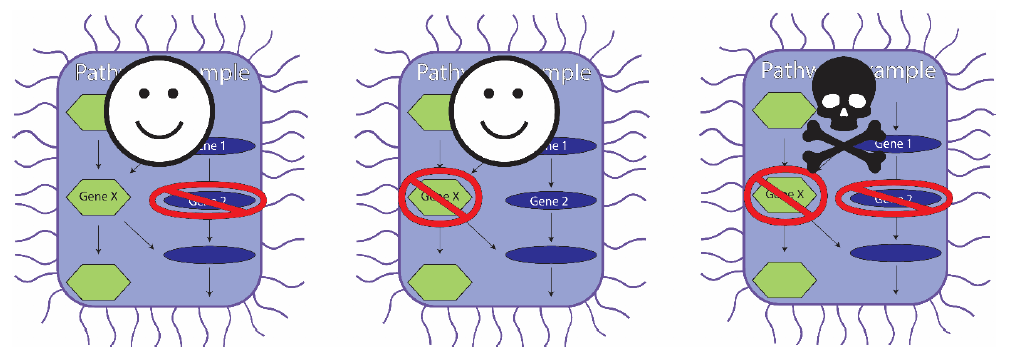
The power of this method can be illustrated in the figure below. If gene 2 (blue pathway) or gene X (green pathway) were switched off alone (the left-hand and middle cases), the cell would not die, because the two genes can substitute for one another. However, in our library, both genes 2 and X will be targeted at the same time in the same cell. Because neither of the two substitutes is present, the pathway cannot synthesise its end-product and the cell dies (at right).
So why do we care? Well, I am part of the Molecular Mycobacteriology Research Unit, and we study Mycobacterium tuberculosis, the bacterium that causes TB. TB is the leading cause of death from infection worldwide and South Africa is particularly badly affected. Current treatment standards requires a minimum 6-month course of multiple antibiotics in combination. This can cause severe side effects and is sometimes not effective at curing the disease. Surprisingly, the choice of combination drugs was made decades ago and arose almost by chance: it didn’t take into account the knowledge we now have about how antibiotics work. To be able to improve chemotherapy for TB, we need new, faster acting antibiotics that have been designed to work together in optimal combinations. Both of these goals require a better understanding not only of how antibiotics work, but of the innermost workings of the bacterium that causes the disease. My library will be built in Mycobacterium smegmatis – a model organism for M. tuberculosis. We aim to figure out which combinations of genes cause cell death when switched off, because we suspect that they might be good antibiotic targets that could improve treatment approaches.

 Written by: Ms Kirsten Winkler
Written by: Ms Kirsten Winkler