The Hans Merensky Chair in Measuring and Modelling Eucalypt Growth & Wood Formation
Understanding growth in the world's most widely planted hardwoods
Update on Rafael’s RNA sequencing experiment
Post authored by Ph.D. candidate Rafael Keret
In an effort to further explore the secondary growth responsiveness of Eucalyptus grandis to xeric environments, an experiment was developed to subject E. grandis juvenile trees to a period of drought.
As part of a greater investigation, the response of the transcriptome to drought will be investigated. The transcriptome is the full complement of messenger-RNA (mRNA) transcripts that is expressed at a particular time and place within a tissue. The tissues of interest for the purposes of our research are that of the cambium and xylem (Figure 1b, c). Understanding the transcriptome dynamics of these tissues during xeric and mesic conditions will enable us to draw parallels between gene expression and the wood properties formed during drought in E. grandis.
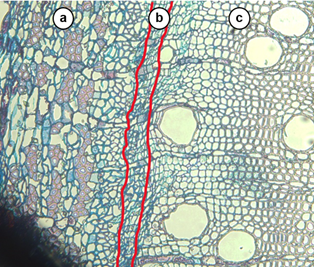 Figure 1: Bright field microscopy cross-section of a E. grandis stem, magnified at the 20x objective. The stem is stained with safranin-Astra blue differential stain. The red borders within the figure divide the regions of the stem into the (a) phloem, (b) cambium, and (c) xylem.
Figure 1: Bright field microscopy cross-section of a E. grandis stem, magnified at the 20x objective. The stem is stained with safranin-Astra blue differential stain. The red borders within the figure divide the regions of the stem into the (a) phloem, (b) cambium, and (c) xylem.
To date, in collaboration with The Institute for Plant Biotechnology (IPB), we have successfully extracted RNA from the stem of E. grandis using the robust CTAB extraction protocol (Figure 2). The resulting RNA was sent to the Central Analytical Facility (CAF) at Stellenbosch University to confirm the RNA integrity, via the use of the TapeStation 4150 system and concentration using the Qubit 4.0 assay.
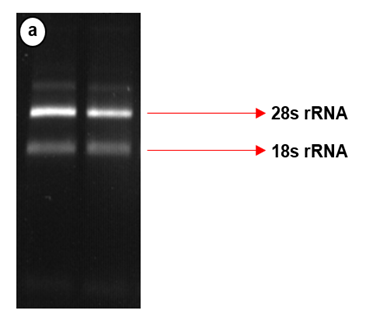 Figure 2: RNA integrity check via the use of a 2% agarose gel. The larger more prominent bands indicate the 28s ribosomal RNA, and the smaller bands (migrated further in the gel) represents the 18s ribosomal RNA.
Figure 2: RNA integrity check via the use of a 2% agarose gel. The larger more prominent bands indicate the 28s ribosomal RNA, and the smaller bands (migrated further in the gel) represents the 18s ribosomal RNA.
Additionally, to test the degree of enrichment of the cambial and xylem tissues (Figure 1 b, c), a polymerase chain reaction (PCR) was conducted on each of the RNA extracts with tissue specific primers (Figure 3). A third primer set, namely that specific to the phloem tissue (Figure 1 a), was added to determine whether this tissue was present in the RNA mixture. It was found that the most highly enriched tissue was the xylem, followed by the cambium and lastly the phloem (almost completely absent) (Figure 3).
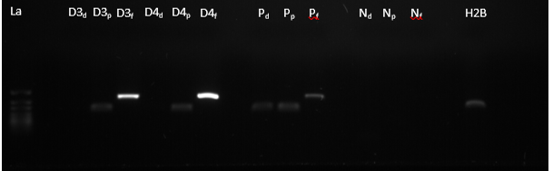 Figure 3: End point GoTaq polymerase chain reaction products, on 2% TBE-agarose gel. La = Ladder, D3d = Droughted 3 phloem specific, D3p = Droughted 3 cambium specific, D3f = Droughted 3 xylem specific, D4d = Droughted 4 phloem specific, D4p = Droughted 4 cambium specific, D4f = Droughted 4 xylem specific, Pd = Positive control phloem specific, Pp = Positive control cambium specific, Pf = Positive control xylem specific, Nd = Negative control phloem specific, Np = Negative control cambium specific, Nf = Negative control xylem specific, H2B = Housekeeping gene.
Figure 3: End point GoTaq polymerase chain reaction products, on 2% TBE-agarose gel. La = Ladder, D3d = Droughted 3 phloem specific, D3p = Droughted 3 cambium specific, D3f = Droughted 3 xylem specific, D4d = Droughted 4 phloem specific, D4p = Droughted 4 cambium specific, D4f = Droughted 4 xylem specific, Pd = Positive control phloem specific, Pp = Positive control cambium specific, Pf = Positive control xylem specific, Nd = Negative control phloem specific, Np = Negative control cambium specific, Nf = Negative control xylem specific, H2B = Housekeeping gene.
Satisfied with the results, the RNA was of a high standard and sent for whole RNA-transcriptome sequencing at Macrogen (South Korea), using the Novaseq-6000 illumina based sequencing platform. However, prior to shipment a buffer was added to the RNA for improved stability during shipment (Figure 4).
 Figure 4: Placement of RNA into buffer for improved stability during shipment, and shipping thereof.
Figure 4: Placement of RNA into buffer for improved stability during shipment, and shipping thereof.
The RNA is now off our hands, and we are eagerly waiting for the data to determine the effects of drought on the transcriptome of E. grandis.